Single-Molecule Studies of the RNA Splicing Process
- Electronic Properties of Molecules Used for Opto-electronic Devices
- Fluorescence Microscopy of Organic Semi-conducting Materials: From Single Molecules to Aggregates
- Single-Molecule Studies of the RNA Splicing Process
- Single-Molecule Studies of Heterogeneous Solvation in Ionic Liquids
- Energy Transfer in DNA-based Dye Arrays
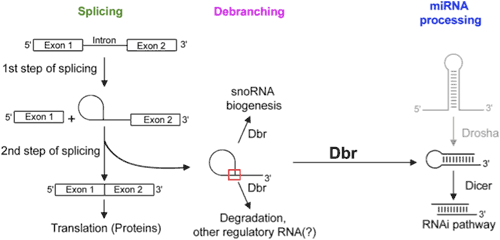
RNA splicing is the process that converts pre-mRNA, which is the initial transcript from DNA, into mRNA, from which protein transcription occurs. Specifically, the protein-coding regions of the pre-mRNA (known as exons) are joined together to form the mature mRNA while the non-coding regions (known as introns) are removed as a lariat structure (see Figure). The lariat contains an unusual 2'-5' phosphodiester linkage. Although, for most mRNAs, the lariat introns are degraded after splicing, some lariat introns have significant roles in important biological processes such as microRNA pathways, snoRNA biogenesis, etc. The lariat debranching enzyme (DBr) plays important role in these biological pathways by linearizing the branched RNA. It does so by selectively cleaving the 2'-5' branched phosphodiester bond of RNA without affecting the normal 3'-5' phosphodiester bond. Although Dbr has striking similarities with other non-specific phosphodiesterase enzymes, it is unique in that it selectively cleaves the 2'-5' phosphodiester bonds of RNA. Therefore it is important to investigate how Dbr recognizes and cleaves its substrate (the 2'-5' phosphodiester bond).
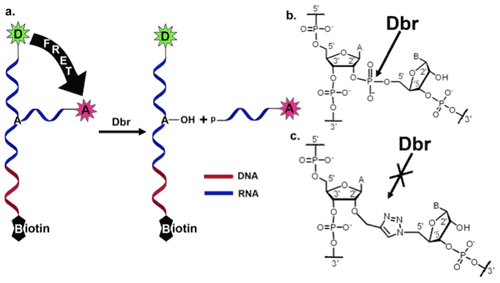
The Das lab has developed a method for solid-phase synthesis of branched RNA containing the 2'-5' phosphodiester bond which mimics the lariat intron. Using this method, it is possible to introduce modifications such as biotin or various fluorescent dyes to the branched RNA during the solid-phase synthesis or through post-synthetic modification. This allows us to follow the debranching of the branched RNA by Dbr at the single-molecule level in the Peteanu lab using Förster Resonance Energy Transfer (FRET) coupled with total internal reflection fluorescence (TIRF) microscopy. For this experiment, the RNA is labeled with a donor dye at the 5'-end of the stem and an acceptor dye at the 3'-end of the branch. Prior to cleavage, energy transfer is possible from the donor to the acceptor. Upon cleavage by Dbr, the acceptor dye is separated from the donor dye, resulting in loss of FRET. The biotin modification at the 3'-end of the stem is used to immobilize the molecules on the surface of a microscope coverslip. Using a non-cleavable substrate analogue (click-branched RNA), we will examine changes in the RNA conformation upon binding with Dbr. The ability to follow the branched RNA and the debranching reaction at the single-molecule level will allow us to probe conformational fluctuations of the branched RNA and the detailed nature of its interaction with Dbr.