Research
Project 1: Recognition of Guanine-Rich DNA and RNA by Homologous PNA
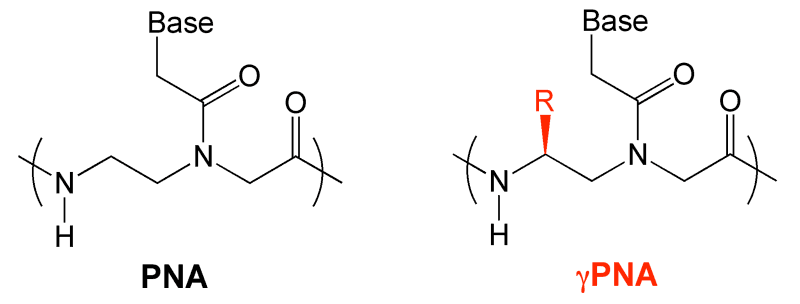 An important component of our research involves peptide nucleic acids (PNAs), which are synthetic molecules that feature the natural hydrogen bonding bases found in DNA (G, A, C and T) attached to an unnatural peptide-like backbone. The bases are spaced apart by approximately the same distance as in DNA or RNA, allowing PNA to hybridize to complementary DNA or RNA targets by the standard Watson-Crick rules of base pairing. The lack of a negative charge on the PNA backbone allows complementary PNA and DNA (or RNA) strands to form more stable double helices than corresponding DNA-DNA, DNA-RNA or RNA-RNA partners, meaning PNA exhibits high affinity for its targets. This raises the possibility of using PNA to interfere with gene expression, based on the hypothesis that strong binding of PNA to an intracellular DNA or RNA target will prevent transcription, splicing, translation or other essential steps in the process by which genetic information is converted into a functional RNA or protein molecule. The unnatural backbone of PNA is also favorable for biological applications, since it is sufficiently distinct from both protein and nucleic acid structures as to be inert toward both protease and nuclease enzymes.
An important component of our research involves peptide nucleic acids (PNAs), which are synthetic molecules that feature the natural hydrogen bonding bases found in DNA (G, A, C and T) attached to an unnatural peptide-like backbone. The bases are spaced apart by approximately the same distance as in DNA or RNA, allowing PNA to hybridize to complementary DNA or RNA targets by the standard Watson-Crick rules of base pairing. The lack of a negative charge on the PNA backbone allows complementary PNA and DNA (or RNA) strands to form more stable double helices than corresponding DNA-DNA, DNA-RNA or RNA-RNA partners, meaning PNA exhibits high affinity for its targets. This raises the possibility of using PNA to interfere with gene expression, based on the hypothesis that strong binding of PNA to an intracellular DNA or RNA target will prevent transcription, splicing, translation or other essential steps in the process by which genetic information is converted into a functional RNA or protein molecule. The unnatural backbone of PNA is also favorable for biological applications, since it is sufficiently distinct from both protein and nucleic acid structures as to be inert toward both protease and nuclease enzymes.
An important breakthrough in the technological development of PNA was the discovery by our collaborator Danith Ly that substituents on the gamma carbon of the PNA backbone can induce additional increases in affinity and improvements in solubility. One aspect of our research is to study the impact of these modifications on the kinetics and thermodynamics of hybridization to complementary DNA and RNA targets.(1) We are also exploiting the high affinity of gPNA to design a new generation of bright fluorescent “miniprobes” that are useful as hybridization probes for nucleic acid detection. More information on gPNA can be found on the Ly group page (insert link here).
A major part of our PNA research over the past decade has focused on studying nontraditional recognition modes. Specifically, we are interested in the guanine tetrad as an alternative to the canonical guanine-cytosine base pair. A G-tetrad can form when four guanines self-assemble via formation of eight hydrogen bonds. The high negative charge density at the core of a G-tetrad by virtue of the four oxygen atoms is normally offset by coordination of a metal ion such as potassium or sodium. Multiple G-tetrads can stack on one another analogous to the stacking of base pairs in a double helix. An assembly of stacked tetrads is referred to as a G-quadruplex.
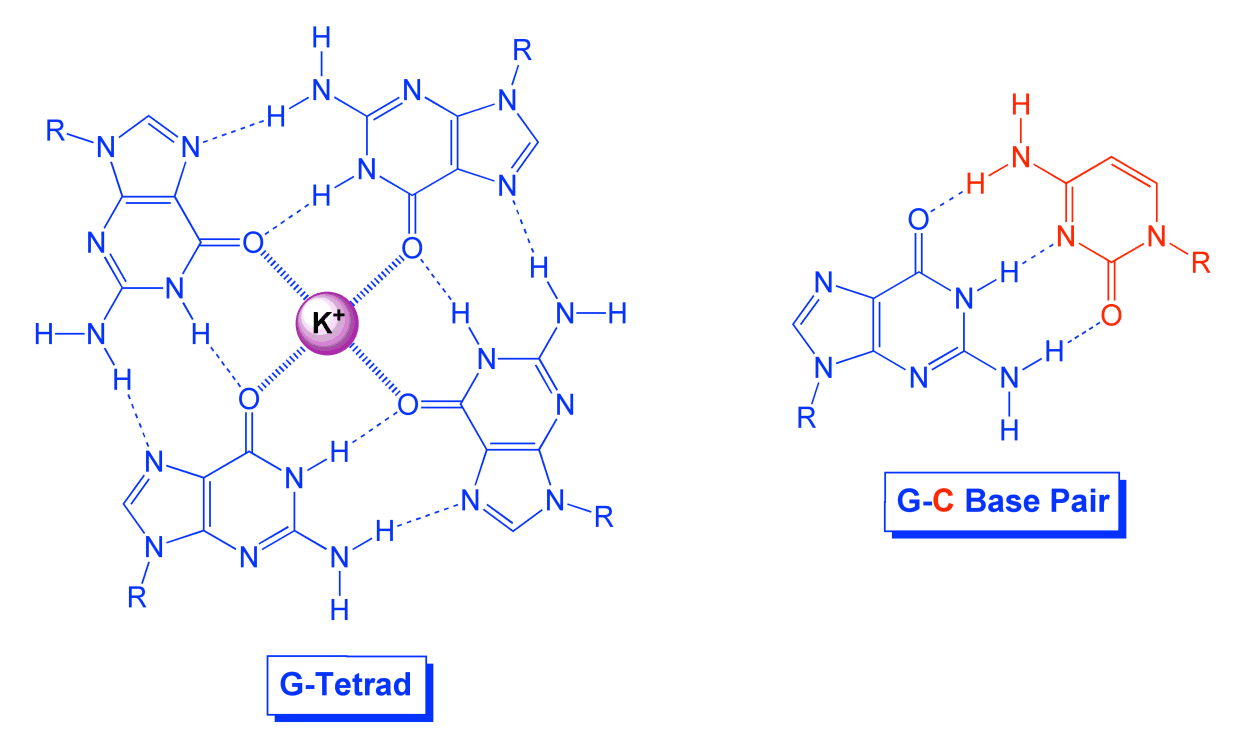
When a single DNA or RNA strand contains a sequence motif represented generally as (G)n-(X)y-(G)n-(X)y-(G)n-(X)y-(G)n, where n ≥ 2 and y ≥ 1, the strand can fold into an intramolecular G-quadruplex featuring n stacked G tetrads connected by (X)y loops. Biological interest in such structures has exploded in the past 10 years due to (a) informatics/sequencing analysis showing that G-quadruplex-forming sequence motifs are more common than would be predicted based solely on statistics, (b) these motifs are over-represented in regions that regulate gene expression (e.g. promoters), particularly in genes that play important roles in cancer, and (c) a growing body of experimental evidence showing that G-quadruplex motifs contribute to regulating gene expression and can be targeted with synthetic compounds.
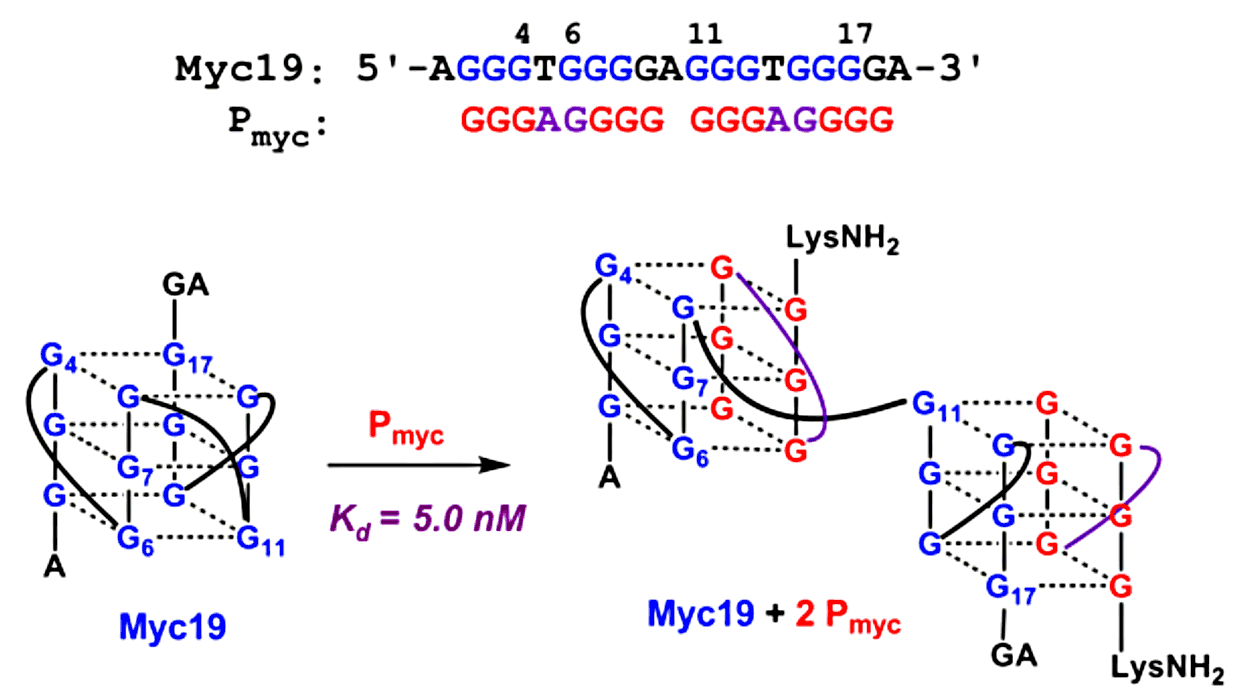
Our work in this area began with a curiosity-driven experiment: would PNA and DNA strands having identical (i.e. homologous) G-rich sequences, recognize one another to form a G-heteroquadruplex? The answer turned out to be “yes”, as demonstrated using homologous PNA and DNA strands having the sequence G4T4G4.(2) We subsequently demonstrated successful hybridization of G-rich PNA to many homologous DNA (3,4) and RNA (5,6) targets, establishing heteroquadruplex formation as a general recognition mode for PNA. For example, the figure below illustrates hybridization of a G-rich PNA 8mer (Pmyc) and a DNA 19mer target (Myc19). The PNA binds in a 2:1 stoichiometry to form adjacent PNA-DNA heteroquadruplexes. The strength of these heteroquadruplexes is quite high, with equilibrium dissociation constants in the low-to-mid nanomolar range. We have explored the use of gamma modifications to improve the selectivity of quadruplex-forming PNAs (7) and our current work, in collaboration with the group of Patty Opresko at the University of Pittsburgh (insert link here) has turned toward establishing the biological implications of quadruplex-forming PNAs, specifically identifying the DNA/RNA targets and effects on gene expression and other properties that result from intracellular delivery of these PNAs.
Publications
- Sahu, B.; Sacui, I.; Rapireddy, S.; Zanotti, K. J.; Bahal, R.; Armitage, B. A.; Ly, D. H. “Synthesis and Characterization of Conformationally Preorganized, (R)-Diethylene Glycol-Containing g-Peptide Nucleic Acids with Superior Hybridization Properties and Water Solubility” J. Org. Chem. 2011, 76, 5614-5627.
- Datta, B.; Schmitt, C.; Armitage, B. A. “Formation of a PNA2-DNA2 Hybrid Quadruplex” J. Am. Chem. Soc. 2003, 125, 4111-4118.
- Roy, S.; Tanious, F. A.; Wilson, W. D.; Ly, D. H.; Armitage, B. A. “High Affinity Homologous Peptide Nucleic Acid Probes for Targeting a Quadruplex Forming Sequence from a MYC Promoter Element” Biochemistry 2007, 46, 10433-10443.
- Roy, S.; Tanious, F. A.; Zanotti, K. J.; Murphy, C. T.; Wilson, W. D.; Ly, D. H.; Armitage, B. A. “Kinetic Discrimination of DNA G Quadruplex Targets by Guanine Rich Peptide Nucleic Acids” Chem. Commun. 2011, 47, 8524-8526.
- Marin, V. L.; Armitage, B. A. “RNA Guanine Quadruplex Invasion by Complementary and Homologous PNA Probes” J. Am. Chem. Soc. 2005, 127, 8032-8033.
- Marin, V. L.; Armitage, B. A. “Hybridization of Complementary and Homologous Peptide Nucleic Acid Oligomers to a Guanine Quadruplex-Forming RNA” Biochemistry 2006, 45, 1745-1754.
- Lusvarghi, S.; Murphy, C. T.; Roy, S.; Tanious, F. A.; Sacui, I.; Wilson, W. D.; Ly, D. H.; Armitage, B. A. “Loop and Backbone Modifications of PNA Improve G Quadruplex Binding Selectivity” J. Am. Chem. Soc. 2009, 131, 18415-18424.