Research
Project 3. Fluoromodules: Dye-Protein Complexes for Fluorescence Imaging
Since the discovery of green fluorescent protein, the use of genetically encodable fluorescent tags for monitoring protein expression, localization and function has become critical for fundamental cell biology and biotechnology. GFP and related proteins that exhibit diverse fluorescence colors and other properties are now widely available and readily expressed as tags on proteins of interest. As part of the Molecular Biosensor and Imaging Center (MBIC), we are developing new fluorescent protein tags that offer alternative colors and improved properties relative to existing proteins.
GFP and its analogues are inherently fluorescent, meaning the fluorophore responsible for absorption and emission of light is covalently integrated within the protein structure. In MBIC’s fluoromodule technology, the fluorophore (e.g. organic dye) is bound non-covalently to the protein. The advantages of this design include high photostability, due to the opportunity to exchange old photobleached fluorophores with new pristine fluorophores, separate optimization of the protein and dye components, and discovery of promiscuous proteins that will bind to many different dyes, leading to a range of colors arising from a single genetically encoded protein tag.
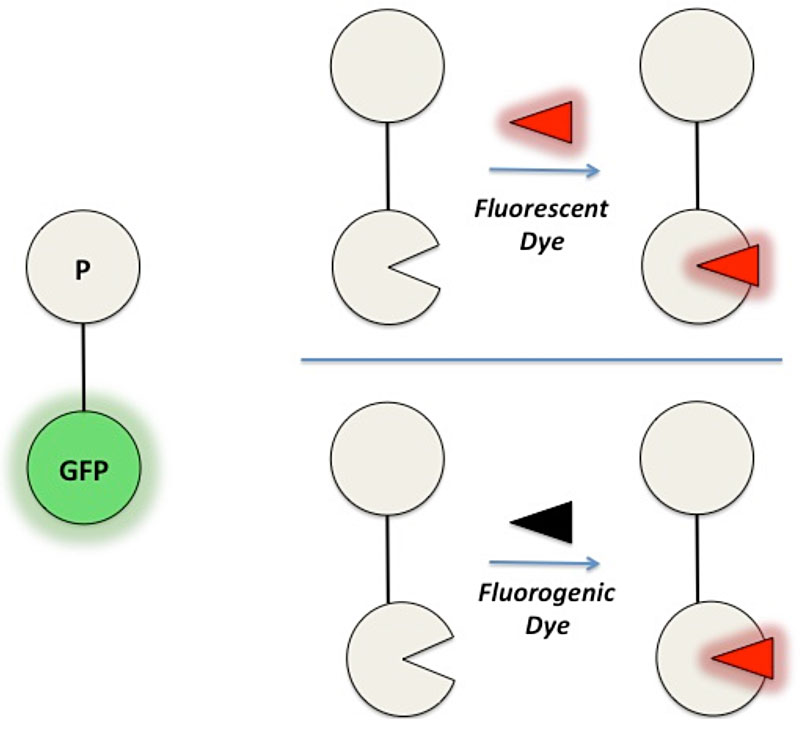
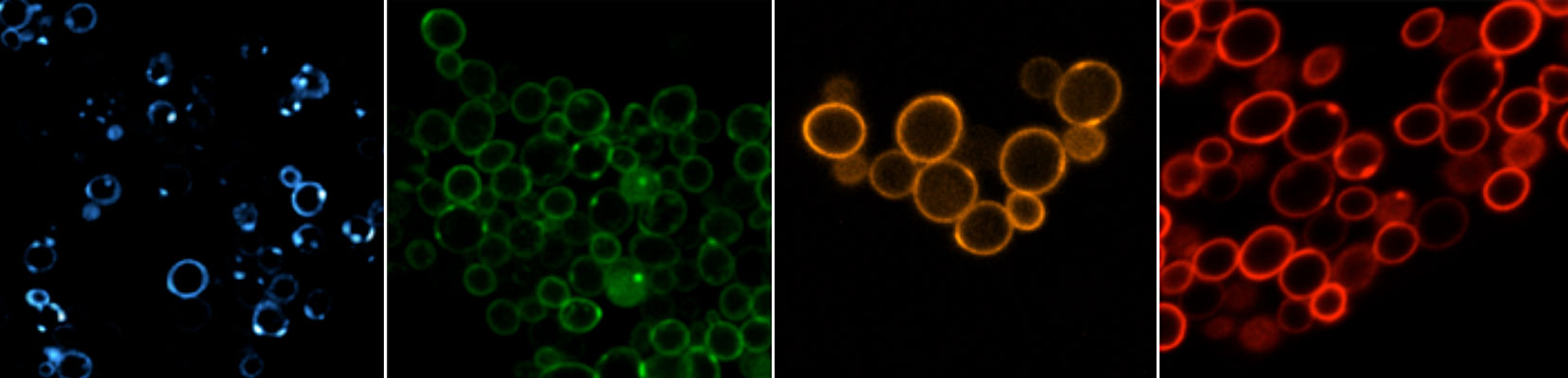
New fluoromodules are obtained from a library of approximately 109 single-chain antibody fragment proteins that are expressed on the surface of yeast cells.1 We begin by adding a fluorogenic dye to the cells. If the dye binds to a protein on the yeast cell surface, it will often exhibit 100-1000-fold enhancements in fluorescence, allowing that particular yeast to be separated from the majority of yeast that do not bind or activate the dye fluorescence. This efficient selection process typically yields proteins that bind their dye targets with low nanomolar equilibrium dissociation constants and generate bright fluorescence.
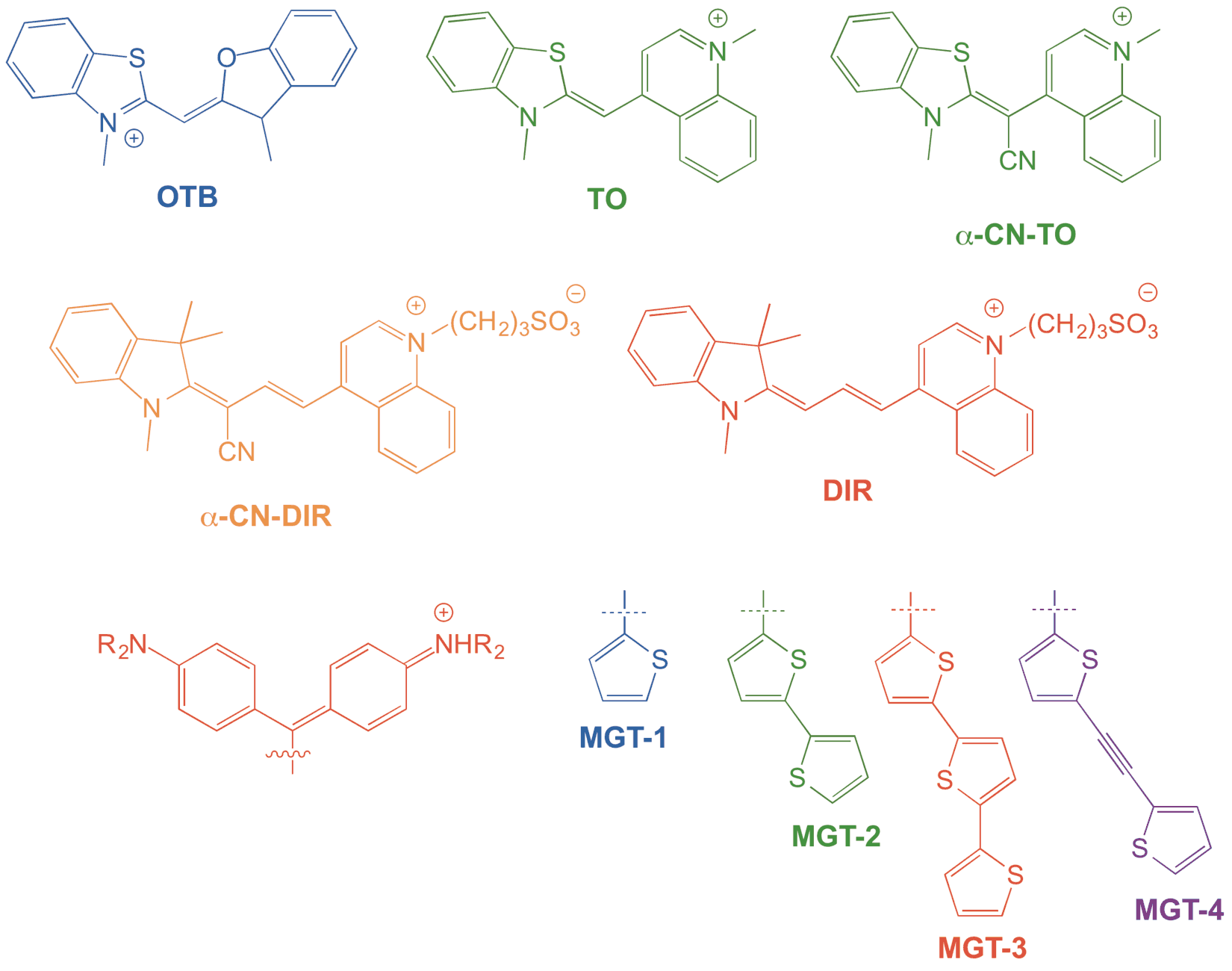
Projects in our lab range from design and synthesis of new fluorogenic dyes to selection and characterization of fluorogen-activating proteins (FAPs). In the past 5 years, we have selected FAPs for the dyes shown below, which cover most of the visible spectrum.2-5 The dyes functionalized with cyano groups exhibit significantly higher photostability than their non-CN counterparts, due in part to suppression of reactivity with singlet oxygen and demonstrating our ability to tune the properties of the fluorogenic dyes independent of the protein. We also collaborate with the Yaron lab to use computational chemistry to understand how the dye structure influences its optical properties and to design new dyes with improved properties.
Publications
(2) Özhalici-Ünal, H.; Lee Pow, C.; Marks, S. A.; Jesper, L. D.; Silva, G. L.; Shank, N. I.; Jones, E. W.; Burnette, J. M., III; Berget, P. B.; Armitage, B. A.: A Rainbow of Fluoromodules: A Promiscuous scFv Protein Binds to and Activates a Diverse Set of Fluorogenic Cyanine Dyes. J. Am. Chem. Soc. 2008, 130, 12620-12621.
(3) Shank, N. I.; Zanotti, K. J.; Lanni, F.; Berget, P. B.; Waggoner, A. S.; Armitage, B. A.: Enhanced Photostability of Genetically Encodable Fluoromodules Based on Fluorogenic Cyanine Dyes and a Promiscuous Protein Partner. J. Am. Chem. Soc. 2009, 131, 12960-12969.
(4) Zanotti, K. J.; Silva, G. L.; Creeger, Y.; Robertson, K. L.; Waggoner, A. S.; Berget, P. B.; Armitage, B. A.: Blue Fluorescent Dye-Protein Complexes Based on Fluorogenic Cyanine Dyes and Single Chain Antibody Fragments. Org. Biomol. Chem. 2011, 9, 1012-1020.
(5) Shank, N. I.; Pham, H. H.; Waggoner, A. S.; Armitage, B. A.: Twisted Cyanines: A Non-Planar Fluorogenic Dye with Superior Photostability and its Use in a Protein-Based Fluoromodule. J. Am. Chem. Soc. 2013, 135, 242-251.