Design of dyes for imaging of biological systems
- Machine Learning for Chemistry
- Photophysics of disordered organic systems
- Design of dyes for imaging of biological systems.
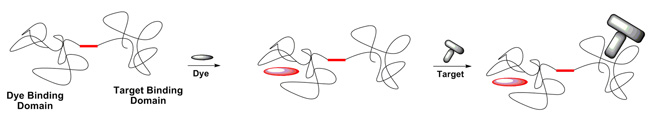
Schematic of an RNA strand used to bind a fluorescent dye to a target protein.
Overview
This work is in collaboration with Bruce Armitage of the Department of Chemistry and Molecular Biosensor and Imaging Center (MBIC) at Carnegie Mellon. The overall goal is to develop fluorogenic dye/RNA aptamer pairs that can be used as components of fluorescent labels and sensors. The above depicts an RNA molecule that consists of two domains, which bind independently to a target of interest and to a fluorogenic dye molecule. Binding of the dye causes it to become fluorescent, meaning that the RNA-dye complex can serve as a fluorescent label for the target. Our goal is to use computation to identify improved dyes.
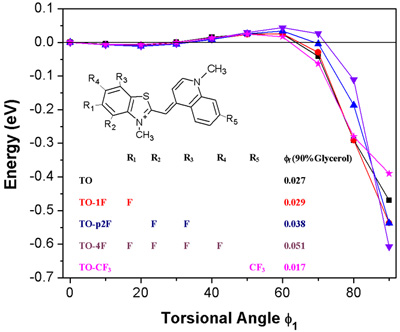
Figure 1: Torsional potential in excited state of fluorogenic dye. In solution, the dye twists beyond 80° and does not emit light. When bound to a target, steric constraints prevent this twist and the dye emits light.
Our recent work illustrates how computation can aid in identifying dyes whose fluorescent properties change on binding to a biomolecule. The above shows the excited state energy as a function of torsional angle between the heterocycles in a series of fluorinated Thiazole Orange (TO) dyes. For all of these dyes, optical excitation is to a nearly planar local minimum with a small barrier leading to a deep twisted minimum. If the molecule overcomes the barrier and reaches the twisted structure, fluorescence is quenched. This occurs only in solution, since binding to a biomolecules hinders the rotation to the twisted structure. Our computed barrier heights are consistent with the trend in observed quantum yields in solution. An optimal dye will rotate, and so quench in solution, yet remain planar when bound to the target molecule.
Our design strategy is to engineering the characteristics of this barrier to enhance and control the degree of fluorescence quenching. In a recent study, we developed an approach to allow computational screening of a library of over a million molecules obtained by placing electron donors and acceptors on a Thiazole Orange framework. We are currently working on control of the barrier through steric effects.
This work is coupled to our general interest on the role of torsional degrees of freedom in organic molecules (see for instance our projects on the effects of disorder on organic semiconductors and on the design of molecular motors).